Our lab focuses on the important basic question of how chromosomes are segregated during cell division to ensure the complete and accurate inheritance of the genome. Centromeres are specified by the incorporation of a histone variant CENP-A, and stable inheritance of this locus is controlled by an epigenetic pathway. Determining the molecular mechanism responsible for the epigenetic inheritance is the major focus of the lab. Chromosome instability is a hallmark of cancer and can drive the formation of tumors. Therefore, how centromere specification is controlled is a basic biological question with great therapeutic potential.
Our long-term goal is to delineate the role of this process in tumorigenesis and translate our basic understanding of the enzymes and proteins involved in this process into therapeutic approaches for targeting proliferative disease. We use a combination of cell biology, biochemical purification, and in vitro reconstitution of centromeric chromatin to discover the mechanism of epigenetic inheritance and CENP-A function during mitosis.
Our long-term goal is to delineate the role of this process in tumorigenesis and translate our basic understanding of the enzymes and proteins involved in this process into therapeutic approaches for targeting proliferative disease. We use a combination of cell biology, biochemical purification, and in vitro reconstitution of centromeric chromatin to discover the mechanism of epigenetic inheritance and CENP-A function during mitosis.
|
Histone Chaperone Networks
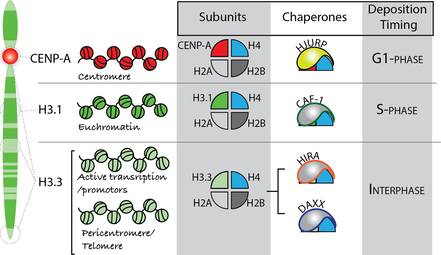
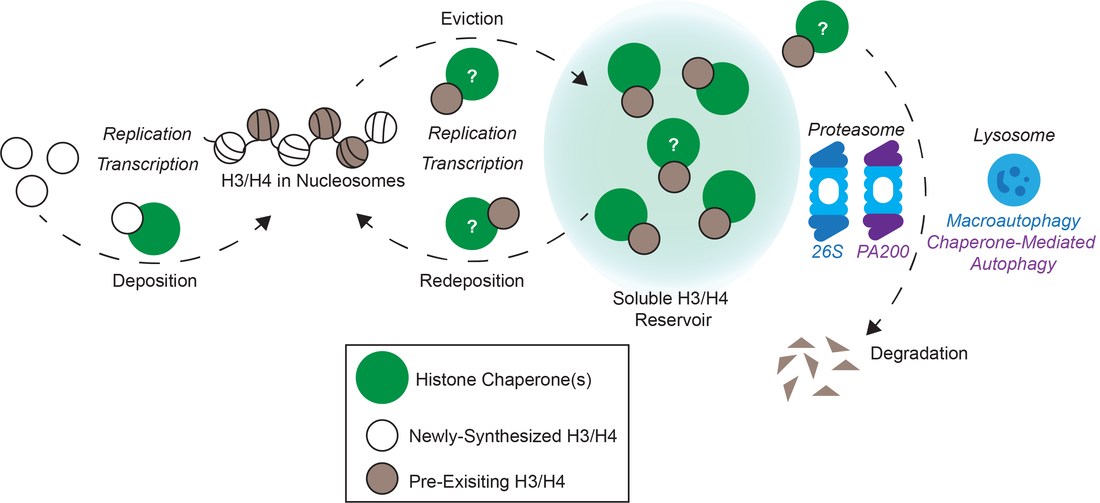
The packing of DNA into histones to form chromatin is a regulated process. A variety of both specific and non-specific histone chaperones shuttle histones to DNA. Chaperones with specificity for histone H3 variants control the unique temporal and spatial distribution of variant-containing nucleosomes within the genome (see figure). The CAF1 histone chaperones supply new H3.1 histones during DNA synthesis. DAXX and HIRA facilitate replication-independent deposition of H3.3 nucleosomes into telomeres, heterochromatin and transcribed regions. And HJURP controls the deposition of CENP-A into nucleosomes specifically at centromeres. Because information in encoded in the posttranslational modification of histones and the placement of histone variants, the retention of histones that are displaced during DNA replication and transcription are key steps in epigenetic inheritance of this information. The stabilization and retention of histones that are evicted during these processes are also controlled by histone chaperones such as NASP (see figure). An excess or a deficit of soluble histones causes replication stress, thus compromising the ability of the cell to faithfully transmit both genetic and epigenetic information across cellular generations. Therefore, the levels of new and existing histones must be tightly regulated by histone chaperones and protein degradation pathways. Our work aims to understand the mechanisms governing the stability, deposition and retention of epigenetic information encoded by histones in human cells. |
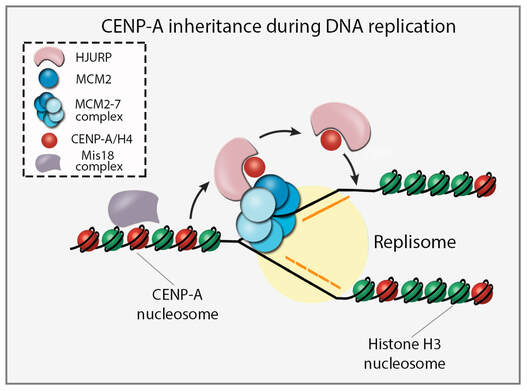
Retention of centromere identity through DNA replication
Centromeric chromatin defines the site of kinetochore formation and ensures faithful chromosome segregation. Centromeric identity is epigenetically specified by the incorporation of CENP-A nucleosomes. DNA replication presents a challenge for inheritance of centromeric identity because nucleosomes are removed to allow for replication fork progression. Despite this challenge, CENP-A nucleosomes are stably retained through S phase. We used BioID to identify proteins transiently associated with CENP-A during DNA replication. We found that during S phase, HJURP transiently associates with centromeres and binds to pre-existing CENP-A, suggesting a distinct role for HJURP in CENP-A retention. We demonstrate that HJURP is required for centromeric nucleosome inheritance during S phase. HJURP co-purifies with the MCM2-7 helicase complex and together with the MCM2 subunit binds CENP-A simultaneously. Therefore, pre-existing CENP-A nucleosomes require an S phase function of the HJURP chaperone and interaction with MCM2 to ensure faithful inheritance of centromere identity through DNA replication.
Centromeric chromatin defines the site of kinetochore formation and ensures faithful chromosome segregation. Centromeric identity is epigenetically specified by the incorporation of CENP-A nucleosomes. DNA replication presents a challenge for inheritance of centromeric identity because nucleosomes are removed to allow for replication fork progression. Despite this challenge, CENP-A nucleosomes are stably retained through S phase. We used BioID to identify proteins transiently associated with CENP-A during DNA replication. We found that during S phase, HJURP transiently associates with centromeres and binds to pre-existing CENP-A, suggesting a distinct role for HJURP in CENP-A retention. We demonstrate that HJURP is required for centromeric nucleosome inheritance during S phase. HJURP co-purifies with the MCM2-7 helicase complex and together with the MCM2 subunit binds CENP-A simultaneously. Therefore, pre-existing CENP-A nucleosomes require an S phase function of the HJURP chaperone and interaction with MCM2 to ensure faithful inheritance of centromere identity through DNA replication.
|
CENP-A Nucleosome Deposition
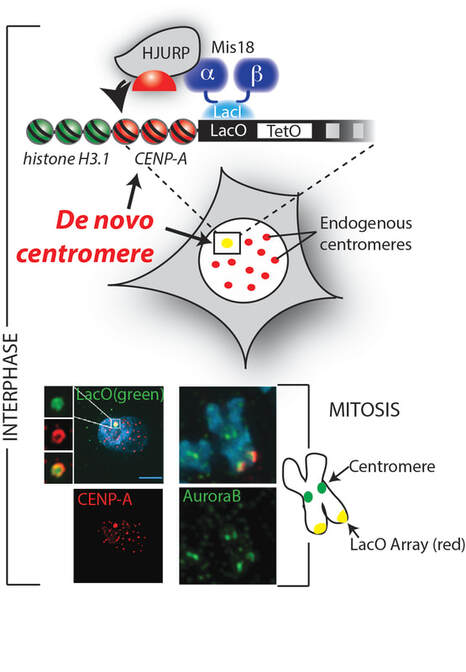
Centromeres are unique chromatin domains that direct the site of kinetochore formation during mitosis and mediate the movement of chromosomes during cell division. Centromeres contain a unique nucleosome in which histone H3 is replaced by centromere protein A (CENP-A). Because of their unique position in chromatin, the CENP-A nucleosome was hypothesized to determine the site of centromere and kinetochore assembly. In order to test the long held assumption that CENP-A dictates the location of the centromere; we developed a novel de novo centromere formation assay, which provides a new and powerful constructive approach to studying centromeres. This system is based on a LacO array that is stably integrated into the long arm of chromosome 1, far away from the existing centromere. Targeting the CENP-A chaperone HJURP or the Mis18 complex to the LacO array, by fusing them to the LacI repressor, drove the stable recruitment of CENP-A nucleosomes to the LacO array at the non-centromeric locus. Ectopically-targeted CENP-A chromatin at the LacO array was sufficient to direct the assembly of a functional centromere as indicated by the recruitment of the constitutive centromere-associated network proteins (CCAN) and formed an active kinetochore during mitosis as indicated by the recruitment of the microtubule-binding protein NDC80, inner centromere protein Aurora A and the formation of stable kinetochore-microtubule attachments. Our work using the de novo centromere assay demonstrates that:
|
|
|
Posttranslational modification of CENP-A
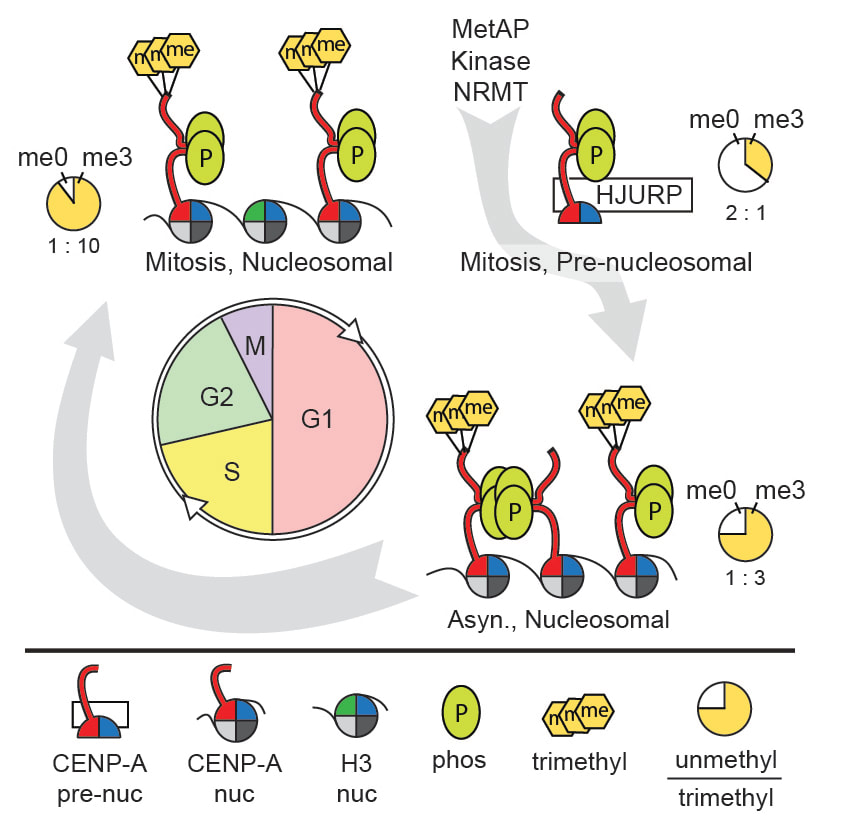
The N-terminal tail of CENP-A is highly divergent from other H3 variants. Canonical histone N-termini are hotspots of conserved post-translational modification; however, no broadly conserved modifications of the vertebrate CENP-A tail have been previously observed. Our lab has identified novel post-translational modifications on human CENP-A N-termini using high-resolution MS. These include the trimethylation of Gly1 at the alpha-amino position and side-chain phosphorylation of Ser16 and Ser18. CENP-A is subjected to constitutive initiating methionine removal, similar to other H3 variants. The nascent N-terminal residue Gly1 becomes trimethylated on the α-amino group. We identified the methyltransferase NRMT as the enzyme responsible for modifying the CENP-A amino terminus. Methylation occurs in the pre-nucleosomal form and marks the majority of CENP-A nucleosomes. Serine 16 and 18 become phosphorylated in pre-nucleosomal CENP-A and are phosphorylated on asynchronous and mitotic nucleosomal CENP-A and is important for chromosome segregation during mitosis. The double phosphorylation motif forms a salt-bridged secondary structure and causes CENP-A N-terminal tails to form intramolecular associations. Analytical ultracentrifugation of phospho-mimetic CENP-A nucleosome arrays demonstrates that phosphorylation results in greater intranucleosome associations and counteracts the hyper-oligomerized state exhibited by unmodified CENP-A nucleosome arrays. Our studies have revealed that the major modifications on the N-terminal tail of CENP-A alter the physical properties of the chromatin fiber at the centromere. We are presently working to understand the function of amino-terminal methylation in CENP-A function. See our review for a full description of how posttranslational modification control centromere function and assembly. Srivastava, S., Zasadzińska, E., and Foltz D.R. (2018) Posttranslational mechanisms controlling centromere function and assembly. Curr Opin Cell Biol. Jun;52:126-135. Srivastava, S. and Foltz D.R. (2018) Posttranslational modifications of CENP-A: marks of distinction (2017) Chromosoma,Sep 127(3):279-290. |